c-MET
OVERVIEW
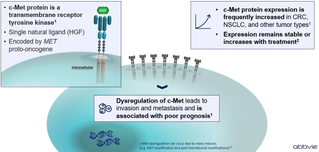
c-Met, also hepatocyte growth factor receptor (HGFR) is a protein encoded by the MET gene. c-Met has tyrosine kinase activity and is expressed on the surface of epithelial and endothelial cells.1
- The only known ligand for c-Met is hepatocyte growth factor (HGF).1
- HGF binding to c-Met activates multiple intracellular signaling pathways including1:
- The MAPK/RAS cascade
- The PI3K/AKT cascade
- The STAT pathway
- These signaling pathways are responsible for driving proliferation, cell survival, migration and invasiveness.1
- c-Met/HGF are essential for embryonic development and tissue repair.2
IMPLICATIONS IN CANCER
Dysregulated MET signaling in cancer occurs through a variety of genetic or epigenetic mechanisms.1-3
- Germline or somatic mutations, chromosomal rearrangement, or MET amplification
- Increased c-Met protein expression
- Increased MET signaling
- Hypoxia-induced overexpression of c-MET
- Alteration of other pathways affecting MET transcriptional activation
- Increased HGF expression
Activation of c-Met directed intracellular signaling pathways is responsible for driving tumor cell proliferation, cell survival, tumor growth, angiogenesis, migration and invasiveness, resistance to therapy, and maintenance of cancer stem cells.1
Notably, alterations in c-Met and/or HGF appear to confer an increased propensity for a more aggressive clinical behavior manifested by invasion, metastasis and resistance to the therapy.1
Oncogenic Expression
c-Met is overexpressed and has prognostic significance in a variety of solid tumors:
- breast cancer (20-30%)4
- non-small cell lung cancer (37-67%; IHC ≥2+)5
- gastric cancer (50%)6
- colorectal cancer (61%)7
- head and neck cancer (up to 80%)8
- kidney cancer (~32%)9
- pancreatic cancer (40-60%)10
Due to the redundant activation of c-Met-directed pathways, c-Met expression on tumors by itself is not a sufficient predictor for activity of small molecule inhibitors.11,12
- The antitumor activity of c-Met inhibitors is generally limited to a subset of patients with tumors that are driven predominately by MET signaling.
- Necessitates selection of patients with MET amplification.
Antibody drug conjugates (ADCs) targeting c-Met represent an attractive therapeutic strategy that does not depend on inhibition of downstream signaling for efficacy but rather on target expression.11,13
- c-Met protein expression is significantly higher in many cancers compared with normal tissues.
Non-Small Cell Lung Cancer
High c-Met expression has been found in NSCLC tumor samples harboring an EGFR mutation.12
- 50% of primary EGFR-TKI-naive NSCLC tumors showed high c-Met expression while only 3% showed c-Met amplification.12
- A significant correlation was found between c-Met expression and EGFR mutations (p = 0.029).12
In advanced-stage NSCLC, MET mutations have a deleterious effect on overall survival (hazard ratio 23.65; p = 0.005) and indicate poor prognosis.14
Related Research
Explore AbbVie's Work
- Garajová I, Giovannetti E, Biasco G, Peters GJ. c-Met as a target for personalized therapy. Transl Oncogenomics. 2015;7(Suppl 1):13-31.
- Sierra JR, Tsao MS. c-MET as a potential therapeutic target and biomarker in cancer. Ther Adv Med Oncol. 2011;3(1 Suppl):S21-S35.
- Koeppen H, Yu W, Zha J, et al. Biomarker analyses from a placebo-controlled phase II study evaluating erlotinib ± onartuzumab in advanced non-small cell lung cancer: MET expression levels are predictive of patient benefit. Clin Cancer Res. 2014;20(17):4488-4498.
- Lengyel E, et al. C-Met overexpression in node-positive breast cancer identifies patients with poor clinical outcome independent of Her2/neu. Int J Cancer. 2005 Feb 10;113(4):678-82.
- Salgia R. MET in Lung Cancer: Biomarker Selection Based on Scientific Rationale. Mol Cancer Ther. 2017;16(4):555-565.
- Mo HN, Liu P. Targeting MET in cancer therapy. Chronic Dis Transl Med. 2017; 3(3): 148–153.
- Liu Y, et al. Prognostic value of c-Met in colorectal cancer: A meta-analysis. World J Gastroenterol. 2015; 21(12): 3706–3710.
- Kim JH, et al. Clinicopathological impacts of high c-Met expression in head and neck squamous cell carcinoma: a meta-analysis and review. Oncotarget. 2017; 8:113120-113128.
- Macher-Goeppinger S, et al. MET expression and copy number status in clear-cell renal cell carcinoma: prognostic value and potential predictive marker. Oncotarget. 2017; 8(1): 1046–1057.
- Kim JH, et al. Prognostic value of c-Met overexpression in pancreatic adenocarcinoma: a meta-analysis. Oncotarget. 2017; 8(42): 73098–73104.
- Wang J, Anderson MG, Oleksijew A, et al. ABBV-399, a c-Met antibody-drug conjugate that targets both MET-amplified and c-Met-overexpressing tumors, irrespective of MET pathway dependence. Clin Cancer Res. 2017;23(4):992-1000.
- Van der Steen N, Deschoolmeester V, Lardon F, et al. cMET in non-small cell lung cancer: pieces of the puzzle. Cancer Res. 2016;76(14 Suppl):Abstract 2242.
- Strickler JH, et al. First-in-Human Phase I, Dose-Escalation and -Expansion Study of Telisotuzumab Vedotin, an Antibody–Drug Conjugate Targeting c-Met, in Patients With Advanced Solid Tumors. J Clin Oncol. 2018;36:3298-3306.
- Lim EH, Zhang SL, Li JL, et al. Using whole genome amplification (WGA) of low-volume biopsies to assess the prognostic role of EGFR, KRAS, p53, and CMET mutations in advanced-stage non-small cell lung cancer (NSCLC). J Thorac Oncol. 2009;4(1):12-21.